Data - an introduction to the world of Pandas
Note: This is an edited version of Cliburn
Chan’s
original tutorial, as part of his Stat-663 course at Duke. All changes
remain licensed as the original, under the terms of the MIT license.
Additionally, sections have been merged from Chris Fonnesbeck’s Pandas
tutorial from the NGCM Summer
Academy,
which are licensed under CC0
terms
(aka ‘public domain’).
Pandas
pandas is a Python package providing fast, flexible, and expressive
data structures designed to work with relational or labeled data
both. It is a fundamental high-level building block for doing practical,
real world data analysis in Python.
pandas is well suited for:
- Tabular data with heterogeneously-typed columns, as you might
find in an SQL table or Excel spreadsheet
- Ordered and unordered (not necessarily fixed-frequency) time
series data.
- Arbitrary matrix data with row and column labels
Virtually any statistical dataset, labeled or unlabeled, can be
converted to a pandas data structure for cleaning, transformation, and
analysis.
Key features
- Easy handling of missing data
- Size mutability: columns can be inserted and deleted from
DataFrame and higher dimensional objects
- Automatic and explicit data alignment: objects can be explicitly
aligned to a set of labels, or the data can be aligned automatically
- Powerful, flexible group by functionality to perform
split-apply-combine operations on data sets
- Intelligent label-based slicing, fancy indexing, and subsetting
of large data sets
- Intuitive merging and joining data sets
- Flexible reshaping and pivoting of data sets
- Hierarchical labeling of axes
- Robust IO tools for loading data from flat files, Excel files,
databases, and HDF5
- Time series functionality: date range generation and frequency
conversion, moving window statistics, moving window linear
regressions, date shifting and lagging, etc.
Working with Series
- A pandas Series is a generationalization of 1d numpy array
- A series has an index that labels each element in the vector.
- A
Series can be thought of as an ordered key-value store.
0 5
1 6
2 7
3 8
4 9
dtype: int64
We can treat Series objects much like numpy vectors
(35, 7.0, 1.5811388300841898)
0 25
1 36
2 49
3 64
4 81
dtype: int64
DataFrames
Inevitably, we want to be able to store, view and manipulate data that
is multivariate, where for every index there are multiple fields or
columns of data, often of varying data type.
A DataFrame is a tabular data structure, encapsulating multiple
series like columns in a spreadsheet. It is directly inspired by the R
DataFrame.
Titanic data
|
survived |
pclass |
sex |
age |
sibsp |
parch |
fare |
embarked |
class |
who |
adult_male |
deck |
embark_town |
alive |
alone |
| 0 |
0 |
3 |
male |
22.0 |
1 |
0 |
7.2500 |
S |
Third |
man |
True |
NaN |
Southampton |
no |
False |
| 1 |
1 |
1 |
female |
38.0 |
1 |
0 |
71.2833 |
C |
First |
woman |
False |
C |
Cherbourg |
yes |
False |
| 2 |
1 |
3 |
female |
26.0 |
0 |
0 |
7.9250 |
S |
Third |
woman |
False |
NaN |
Southampton |
yes |
True |
| 3 |
1 |
1 |
female |
35.0 |
1 |
0 |
53.1000 |
S |
First |
woman |
False |
C |
Southampton |
yes |
False |
| 4 |
0 |
3 |
male |
35.0 |
0 |
0 |
8.0500 |
S |
Third |
man |
True |
NaN |
Southampton |
no |
True |
Index(['survived', 'pclass', 'sex', 'age', 'sibsp', 'parch', 'fare',
'embarked', 'class', 'who', 'adult_male', 'deck', 'embark_town',
'alive', 'alone'],
dtype='object')
survived int64
sex object
age float64
fare float64
embarked object
class object
who object
deck object
embark_town object
dtype: object
Summarizing a data frame
|
survived |
age |
fare |
| count |
891.000000 |
714.000000 |
891.000000 |
| mean |
0.383838 |
29.699118 |
32.204208 |
| std |
0.486592 |
14.526497 |
49.693429 |
| min |
0.000000 |
0.420000 |
0.000000 |
| 25% |
0.000000 |
20.125000 |
7.910400 |
| 50% |
0.000000 |
28.000000 |
14.454200 |
| 75% |
1.000000 |
38.000000 |
31.000000 |
| max |
1.000000 |
80.000000 |
512.329200 |
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
NaN |
Southampton |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
| 2 |
1 |
female |
26.0 |
7.9250 |
S |
Third |
woman |
NaN |
Southampton |
| 3 |
1 |
female |
35.0 |
53.1000 |
S |
First |
woman |
C |
Southampton |
| 4 |
0 |
male |
35.0 |
8.0500 |
S |
Third |
man |
NaN |
Southampton |
| 5 |
0 |
male |
NaN |
8.4583 |
Q |
Third |
man |
NaN |
Queenstown |
| 6 |
0 |
male |
54.0 |
51.8625 |
S |
First |
man |
E |
Southampton |
| 7 |
0 |
male |
2.0 |
21.0750 |
S |
Third |
child |
NaN |
Southampton |
| 8 |
1 |
female |
27.0 |
11.1333 |
S |
Third |
woman |
NaN |
Southampton |
| 9 |
1 |
female |
14.0 |
30.0708 |
C |
Second |
child |
NaN |
Cherbourg |
| 10 |
1 |
female |
4.0 |
16.7000 |
S |
Third |
child |
G |
Southampton |
| 11 |
1 |
female |
58.0 |
26.5500 |
S |
First |
woman |
C |
Southampton |
| 12 |
0 |
male |
20.0 |
8.0500 |
S |
Third |
man |
NaN |
Southampton |
| 13 |
0 |
male |
39.0 |
31.2750 |
S |
Third |
man |
NaN |
Southampton |
| 14 |
0 |
female |
14.0 |
7.8542 |
S |
Third |
child |
NaN |
Southampton |
| 15 |
1 |
female |
55.0 |
16.0000 |
S |
Second |
woman |
NaN |
Southampton |
| 16 |
0 |
male |
2.0 |
29.1250 |
Q |
Third |
child |
NaN |
Queenstown |
| 17 |
1 |
male |
NaN |
13.0000 |
S |
Second |
man |
NaN |
Southampton |
| 18 |
0 |
female |
31.0 |
18.0000 |
S |
Third |
woman |
NaN |
Southampton |
| 19 |
1 |
female |
NaN |
7.2250 |
C |
Third |
woman |
NaN |
Cherbourg |
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
| 886 |
0 |
male |
27.0 |
13.00 |
S |
Second |
man |
NaN |
Southampton |
| 887 |
1 |
female |
19.0 |
30.00 |
S |
First |
woman |
B |
Southampton |
| 888 |
0 |
female |
NaN |
23.45 |
S |
Third |
woman |
NaN |
Southampton |
| 889 |
1 |
male |
26.0 |
30.00 |
C |
First |
man |
C |
Cherbourg |
| 890 |
0 |
male |
32.0 |
7.75 |
Q |
Third |
man |
NaN |
Queenstown |
Index(['survived', 'sex', 'age', 'fare', 'embarked', 'class', 'who', 'deck',
'embark_town'],
dtype='object')
RangeIndex(start=0, stop=891, step=1)
Indexing
The default indexing mode for dataframes with df[X] is to access the
DataFrame’s columns:
|
sex |
age |
class |
| 0 |
male |
22.0 |
Third |
| 1 |
female |
38.0 |
First |
| 2 |
female |
26.0 |
Third |
| 3 |
female |
35.0 |
First |
| 4 |
male |
35.0 |
Third |
Using the iloc helper for indexing
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
NaN |
Southampton |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
| 2 |
1 |
female |
26.0 |
7.9250 |
S |
Third |
woman |
NaN |
Southampton |
<pandas.core.indexing._iLocIndexer at 0x106cecb70>
survived 0
sex male
age 22
fare 7.25
embarked S
class Third
who man
deck NaN
embark_town Southampton
Name: 0, dtype: object
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
NaN |
Southampton |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
| 2 |
1 |
female |
26.0 |
7.9250 |
S |
Third |
woman |
NaN |
Southampton |
| 3 |
1 |
female |
35.0 |
53.1000 |
S |
First |
woman |
C |
Southampton |
| 4 |
0 |
male |
35.0 |
8.0500 |
S |
Third |
man |
NaN |
Southampton |
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
NaN |
Southampton |
| 10 |
1 |
female |
4.0 |
16.7000 |
S |
Third |
child |
G |
Southampton |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
| 5 |
0 |
male |
NaN |
8.4583 |
Q |
Third |
man |
NaN |
Queenstown |
10 4.0
11 58.0
12 20.0
13 39.0
14 14.0
Name: age, dtype: float64
|
age |
| 10 |
4.0 |
| 11 |
58.0 |
| 12 |
20.0 |
| 13 |
39.0 |
| 14 |
14.0 |
File "<ipython-input-47-0c88d841b460>", line 1
titanic[titanic. < 2]
^
SyntaxError: invalid syntax
0 0
1 0
2 0
3 0
4 0
5 0
6 0
7 0
8 0
9 0
Name: new column, dtype: int64
Int64Index([78, 164, 172, 183, 305, 381, 386, 469, 644, 755, 788, 803, 827,
831],
dtype='int64')
|
name |
age |
| id |
|
|
| 123 |
Alice |
20 |
| 989 |
Bob |
30 |
.iloc vs .loc
These are two accessors with a key difference:
.iloc indexes positionally.loc indexes by label
name Alice
age 20
Name: 123, dtype: object
name Alice
age 20
Name: 123, dtype: object
Sorting and ordering data
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
NaN |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
| 2 |
1 |
female |
26.0 |
7.9250 |
S |
Third |
woman |
NaN |
Southampton |
0 |
| 3 |
1 |
female |
35.0 |
53.1000 |
S |
First |
woman |
C |
Southampton |
0 |
| 4 |
0 |
male |
35.0 |
8.0500 |
S |
Third |
man |
NaN |
Southampton |
0 |
The sort_index method is designed to sort a DataFrame by either its
index or its columns:
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 890 |
0 |
male |
32.0 |
7.75 |
Q |
Third |
man |
NaN |
Queenstown |
0 |
| 889 |
1 |
male |
26.0 |
30.00 |
C |
First |
man |
C |
Cherbourg |
0 |
| 888 |
0 |
female |
NaN |
23.45 |
S |
Third |
woman |
NaN |
Southampton |
0 |
| 887 |
1 |
female |
19.0 |
30.00 |
S |
First |
woman |
B |
Southampton |
0 |
| 886 |
0 |
male |
27.0 |
13.00 |
S |
Second |
man |
NaN |
Southampton |
0 |
Since the Titanic index is already sorted, it’s easier to illustrate how
to use it for the index with a small test DF:
|
A |
| 100 |
1 |
| 29 |
2 |
| 234 |
3 |
| 1 |
4 |
| 150 |
5 |
|
A |
| 1 |
4 |
| 29 |
2 |
| 100 |
1 |
| 150 |
5 |
| 234 |
3 |
Pandas also makes it easy to sort on the values of the DF:
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 803 |
1 |
male |
0.42 |
8.5167 |
C |
Third |
child |
NaN |
Cherbourg |
0 |
| 755 |
1 |
male |
0.67 |
14.5000 |
S |
Second |
child |
NaN |
Southampton |
0 |
| 644 |
1 |
female |
0.75 |
19.2583 |
C |
Third |
child |
NaN |
Cherbourg |
0 |
| 469 |
1 |
female |
0.75 |
19.2583 |
C |
Third |
child |
NaN |
Cherbourg |
0 |
| 78 |
1 |
male |
0.83 |
29.0000 |
S |
Second |
child |
NaN |
Southampton |
0 |
And we can sort on more than one column in a single call:
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 164 |
0 |
male |
1.0 |
39.6875 |
S |
Third |
child |
NaN |
Southampton |
0 |
| 386 |
0 |
male |
1.0 |
46.9000 |
S |
Third |
child |
NaN |
Southampton |
0 |
| 7 |
0 |
male |
2.0 |
21.0750 |
S |
Third |
child |
NaN |
Southampton |
0 |
| 16 |
0 |
male |
2.0 |
29.1250 |
Q |
Third |
child |
NaN |
Queenstown |
0 |
| 119 |
0 |
female |
2.0 |
31.2750 |
S |
Third |
child |
NaN |
Southampton |
0 |
Note: both the index and the columns can be named:
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| id |
|
|
|
|
|
|
|
|
|
|
| 851 |
0 |
male |
74.0 |
7.7750 |
S |
Third |
man |
NaN |
Southampton |
0 |
| 96 |
0 |
male |
71.0 |
34.6542 |
C |
First |
man |
A |
Cherbourg |
0 |
| 493 |
0 |
male |
71.0 |
49.5042 |
C |
First |
man |
NaN |
Cherbourg |
0 |
| 116 |
0 |
male |
70.5 |
7.7500 |
Q |
Third |
man |
NaN |
Queenstown |
0 |
| 672 |
0 |
male |
70.0 |
10.5000 |
S |
Second |
man |
NaN |
Southampton |
0 |
Grouping data
What is a GroubBy object?
<pandas.core.groupby.DataFrameGroupBy object at 0x101c3b860>
name: ('female', 'First')
group:
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
| 3 |
1 |
female |
35.0 |
53.1000 |
S |
First |
woman |
C |
Southampton |
0 |
name: ('female', 'Second')
group:
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 9 |
1 |
female |
14.0 |
30.0708 |
C |
Second |
child |
NaN |
Cherbourg |
0 |
| 15 |
1 |
female |
55.0 |
16.0000 |
S |
Second |
woman |
NaN |
Southampton |
0 |
name: ('female', 'Third')
group:
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 2 |
1 |
female |
26.0 |
7.9250 |
S |
Third |
woman |
NaN |
Southampton |
0 |
| 8 |
1 |
female |
27.0 |
11.1333 |
S |
Third |
woman |
NaN |
Southampton |
0 |
name: ('male', 'First')
group:
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 6 |
0 |
male |
54.0 |
51.8625 |
S |
First |
man |
E |
Southampton |
0 |
| 23 |
1 |
male |
28.0 |
35.5000 |
S |
First |
man |
A |
Southampton |
0 |
name: ('male', 'Second')
group:
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 17 |
1 |
male |
NaN |
13.0 |
S |
Second |
man |
NaN |
Southampton |
0 |
| 20 |
0 |
male |
35.0 |
26.0 |
S |
Second |
man |
NaN |
Southampton |
0 |
name: ('male', 'Third')
group:
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.25 |
S |
Third |
man |
NaN |
Southampton |
0 |
| 4 |
0 |
male |
35.0 |
8.05 |
S |
Third |
man |
NaN |
Southampton |
0 |
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 9 |
1 |
female |
14.0 |
30.0708 |
C |
Second |
child |
NaN |
Cherbourg |
0 |
| 15 |
1 |
female |
55.0 |
16.0000 |
S |
Second |
woman |
NaN |
Southampton |
0 |
| 41 |
0 |
female |
27.0 |
21.0000 |
S |
Second |
woman |
NaN |
Southampton |
0 |
| 43 |
1 |
female |
3.0 |
41.5792 |
C |
Second |
child |
NaN |
Cherbourg |
0 |
| 53 |
1 |
female |
29.0 |
26.0000 |
S |
Second |
woman |
NaN |
Southampton |
0 |
The GroubBy object has a number of aggregation methods that will then
compute summary statistics over the group members, e.g.:
|
attributes |
survived |
age |
fare |
embarked |
who |
deck |
embark_town |
new column |
| sex |
class |
|
|
|
|
|
|
|
|
| female |
First |
94 |
85 |
94 |
92 |
94 |
81 |
92 |
94 |
| Second |
76 |
74 |
76 |
76 |
76 |
10 |
76 |
76 |
| Third |
144 |
102 |
144 |
144 |
144 |
6 |
144 |
144 |
| male |
First |
122 |
101 |
122 |
122 |
122 |
94 |
122 |
122 |
| Second |
108 |
99 |
108 |
108 |
108 |
6 |
108 |
108 |
| Third |
347 |
253 |
347 |
347 |
347 |
6 |
347 |
347 |
Why Kate Winslett survived and Leonardo DiCaprio didn’t
|
attributes |
survived |
| sex |
class |
|
| female |
First |
0.968085 |
| Second |
0.921053 |
| Third |
0.500000 |
| male |
First |
0.368852 |
| Second |
0.157407 |
| Third |
0.135447 |
Of the females who were in first class, count the number from each embarking town
| attributes |
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
new column |
| embark_town |
|
|
|
|
|
|
|
|
|
| Cherbourg |
43 |
43 |
38 |
43 |
43 |
43 |
43 |
35 |
43 |
| Queenstown |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
| Southampton |
48 |
48 |
44 |
48 |
48 |
48 |
48 |
43 |
48 |
Since count counts non-missing data, we’re really interested in the
maximum value for each row, which we can obtain directly:
embark_town
Cherbourg 43
Queenstown 1
Southampton 48
dtype: int64
Cross-tabulation
| class |
First |
Second |
Third |
| survived |
|
|
|
| 0 |
80 |
97 |
372 |
| 1 |
136 |
87 |
119 |
We can also get multiple summaries at the same time
The agg method is the most flexible, as it allows us to specify
directly which functions we want to call, and where:
|
embarked |
age |
survived |
|
count |
mean |
median |
my_func |
sum |
| embark_town |
|
|
|
|
|
| Cherbourg |
43 |
36.052632 |
37.0 |
60.0 |
42 |
| Queenstown |
1 |
33.000000 |
33.0 |
33.0 |
1 |
| Southampton |
48 |
32.704545 |
33.0 |
63.0 |
46 |
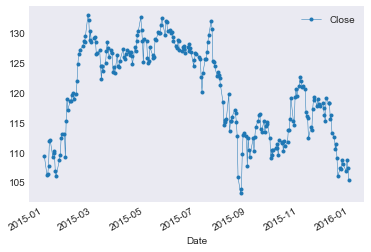
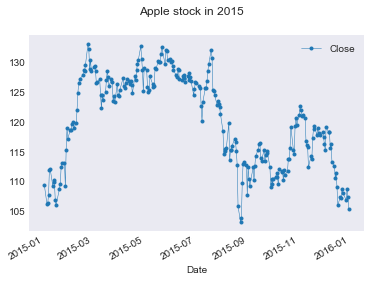
Making plots with pandas
Note: you may need to run
pip install pandas-datareader
to install the specialized readers.
|
Open |
High |
Low |
Close |
Adj Close |
Volume |
| Date |
|
|
|
|
|
|
| 2015-01-02 |
111.389999 |
111.440002 |
107.349998 |
109.330002 |
103.866470 |
53204600 |
| 2015-01-05 |
108.290001 |
108.650002 |
105.410004 |
106.250000 |
100.940392 |
64285500 |
| 2015-01-06 |
106.540001 |
107.430000 |
104.629997 |
106.260002 |
100.949890 |
65797100 |
| 2015-01-07 |
107.199997 |
108.199997 |
106.699997 |
107.750000 |
102.365440 |
40105900 |
| 2015-01-08 |
109.230003 |
112.150002 |
108.699997 |
111.889999 |
106.298531 |
59364500 |
|
Open |
High |
Low |
Close |
Adj Close |
Volume |
| Date |
|
|
|
|
|
|
| 2015-12-24 |
109.000000 |
109.000000 |
107.949997 |
108.029999 |
104.380112 |
13570400 |
| 2015-12-28 |
107.589996 |
107.690002 |
106.180000 |
106.820000 |
103.210999 |
26704200 |
| 2015-12-29 |
106.959999 |
109.430000 |
106.860001 |
108.739998 |
105.066116 |
30931200 |
| 2015-12-30 |
108.580002 |
108.699997 |
107.180000 |
107.320000 |
103.694107 |
25213800 |
| 2015-12-31 |
107.010002 |
107.029999 |
104.820000 |
105.260002 |
101.703697 |
40635300 |
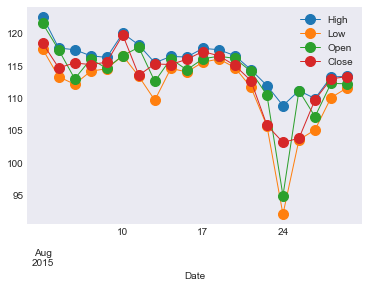
Let’s save this data to a CSV file so we don’t need to re-download it on
every run:
|
Open |
High |
Low |
Close |
Adj Close |
Volume |
| Date |
|
|
|
|
|
|
| 2015-08-03 |
121.500000 |
122.570000 |
117.519997 |
118.440002 |
113.437157 |
69976000 |
| 2015-08-04 |
117.419998 |
117.699997 |
113.250000 |
114.639999 |
109.797668 |
124138600 |
| 2015-08-05 |
112.949997 |
117.440002 |
112.099998 |
115.400002 |
110.525574 |
99312600 |
| 2015-08-06 |
115.970001 |
116.500000 |
114.120003 |
115.129997 |
110.766090 |
52903000 |
| 2015-08-07 |
114.580002 |
116.250000 |
114.500000 |
115.519997 |
111.141319 |
38670400 |
/Users/fperez/usr/conda/envs/s159/lib/python3.6/site-packages/pandas/plotting/_core.py:1714: UserWarning: Pandas doesn't allow columns to be created via a new attribute name - see https://pandas.pydata.org/pandas-docs/stable/indexing.html#attribute-access
series.name = label
Data conversions
One of the nicest features of pandas is the ease of converting
tabular data across different storage formats. We will illustrate by
converting the titanic dataframe into multiple formats.
CSV
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
NaN |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
Excel
You may need to first install openpyxl:
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
NaN |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
Relational Database
/Users/fperez/usr/conda/envs/s159/lib/python3.6/site-packages/pandas/core/generic.py:1534: UserWarning: The spaces in these column names will not be changed. In pandas versions < 0.14, spaces were converted to underscores.
chunksize=chunksize, dtype=dtype)
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
None |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
JSON
|
age |
class |
deck |
embark_town |
embarked |
fare |
new column |
sex |
survived |
who |
| 0 |
22.0 |
Third |
None |
Southampton |
S |
7.2500 |
0 |
male |
0 |
man |
| 1 |
38.0 |
First |
C |
Cherbourg |
C |
71.2833 |
0 |
female |
1 |
woman |
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
None |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
HDF5
The HDF5
format
was designed in the Earth Sciences community but it can be an excellent
general purpose tool. It’s efficient and type-safe, so you can store
complex dataframes in it and recover them back without information loss,
using the to_hdf method:
/Users/fperez/usr/conda/envs/s159/lib/python3.6/site-packages/pandas/core/generic.py:1471: PerformanceWarning:
your performance may suffer as PyTables will pickle object types that it cannot
map directly to c-types [inferred_type->mixed,key->block1_values] [items->['sex', 'embarked', 'class', 'who', 'deck', 'embark_town']]
return pytables.to_hdf(path_or_buf, key, self, **kwargs)
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
None |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
Feather
You may need to install the
Feather
support first:
conda install -c conda-forge feather-format
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
None |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
| 2 |
1 |
female |
4.0 |
16.7000 |
S |
Third |
child |
G |
Southampton |
0 |
| 3 |
0 |
female |
28.0 |
7.8958 |
S |
Third |
woman |
None |
Southampton |
0 |
| 4 |
0 |
male |
NaN |
7.8958 |
S |
Third |
man |
None |
Southampton |
0 |
|
survived |
sex |
age |
fare |
embarked |
class |
who |
deck |
embark_town |
new column |
| 0 |
0 |
male |
22.0 |
7.2500 |
S |
Third |
man |
None |
Southampton |
0 |
| 1 |
1 |
female |
38.0 |
71.2833 |
C |
First |
woman |
C |
Cherbourg |
0 |
| 2 |
1 |
female |
4.0 |
16.7000 |
S |
Third |
child |
G |
Southampton |
0 |
| 3 |
0 |
female |
28.0 |
7.8958 |
S |
Third |
woman |
None |
Southampton |
0 |
| 4 |
0 |
male |
NaN |
7.8958 |
S |
Third |
man |
None |
Southampton |
0 |